Into the Unknown of Cell Biology: Fluorescent Protein Technology in Combination with Yeast Genetic Engineering Reveals Unprecedented Roles of Metabolic Enzymes.
Tel: 66 (0) 2441-9003 – 7 Ext. 1274
Email: chalongrat.nor mahidol.edu
mahidol.edu
Ph.D. (Biology), University of California, San Diego, 2013
Academic Program(s)
Molecular Genetics and Genetic Engineering
Molecular and Integrative Biosciences
Our research team focuses on a fascinating aspect of cell biology: how metabolic enzymes assemble into large, organized structures. We’ve found that many of these enzymes don’t just exist as individual proteins but can come together to form visible filaments and other complex shapes. Our work has shown that this enzyme polymerization is a key way a cell can regulate its metabolism.
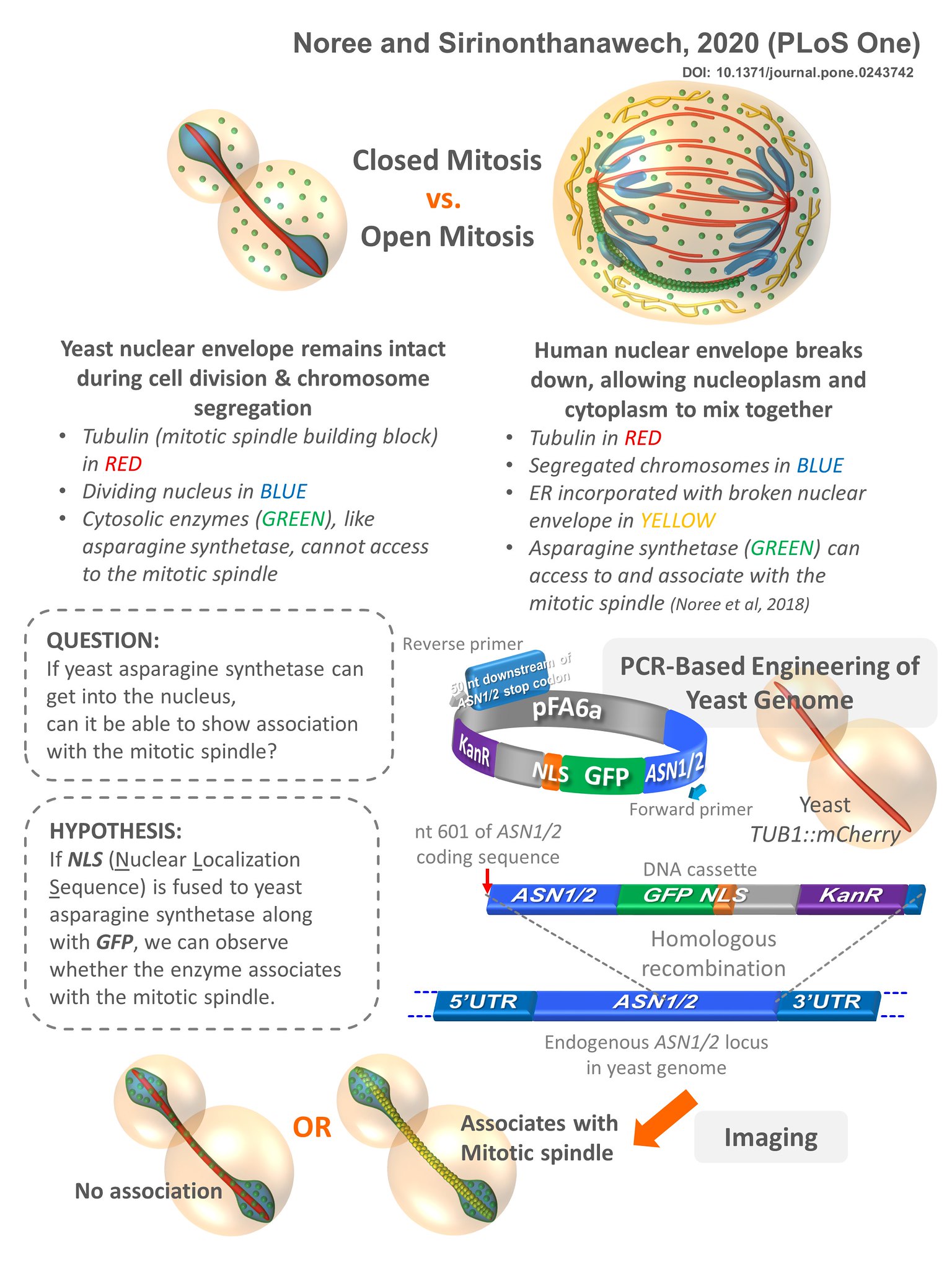
To illustrate this, we’ve extensively studied the yeast enzyme asparagine synthetase. Our research has shown how it forms these structures in response to nutrient stress, directly impacting its activity. Apart from this primary metabolic role, we’ve also investigated a notable moonlighting function of asparagine synthetase in human cells, where it associates with the mitotic spindle, suggesting a potential role in cell division. Using yeast as a model system, we confirmed that the catalytic activity of asparagine synthetase is not required for this function. By introducing a mutation known to disrupt its activity into the yeast genome, we found that its ability to associate with the mitotic spindle remained intact.
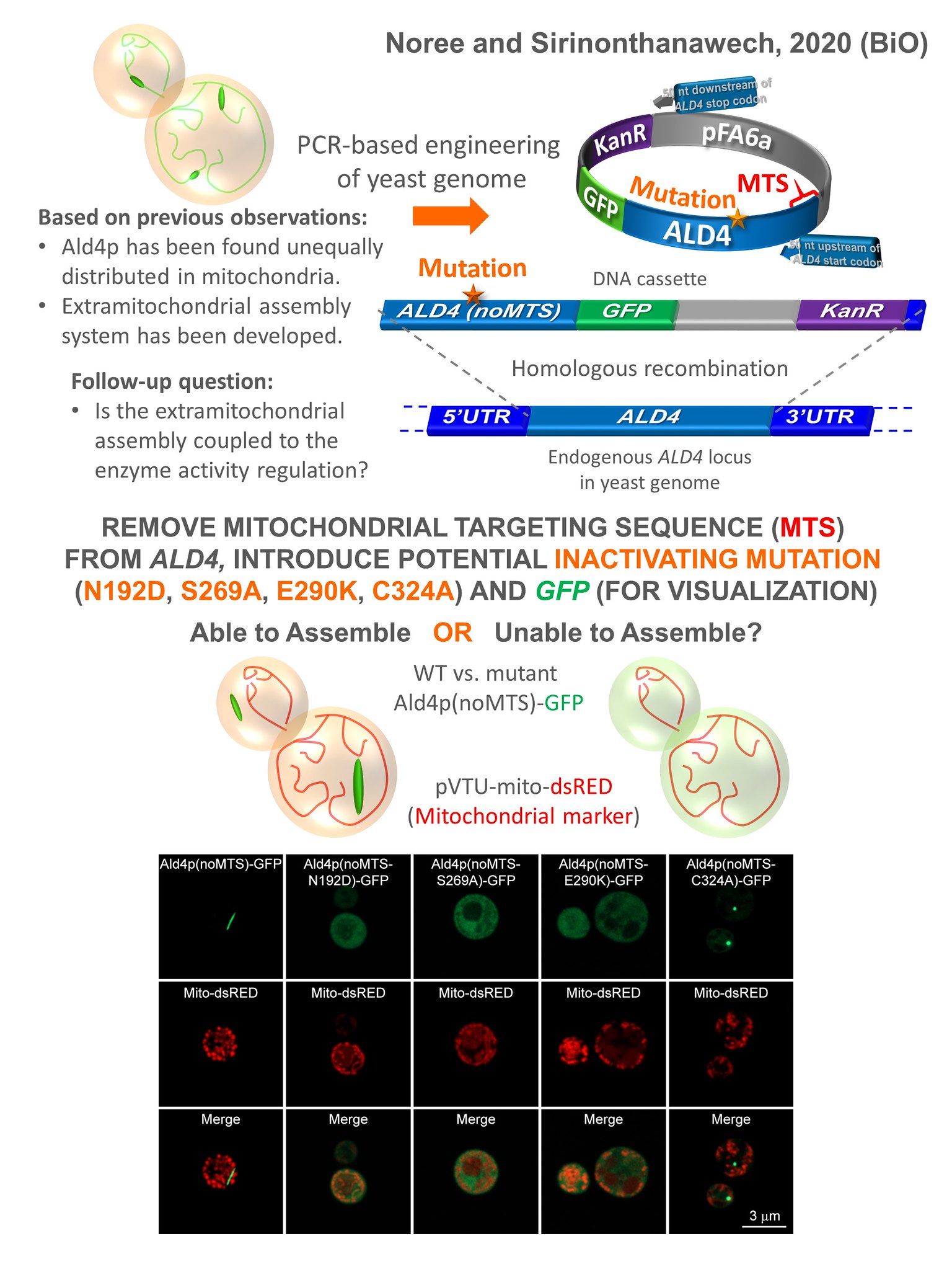
Beyond cytosolic enzymes, we have recently expanded our work to include mitochondrial enzymes. We’ve discovered that a major mitochondrial enzyme, aldehyde dehydrogenase (Ald4p), can have a portion of its population form gigantic supramolecular structures in the cytoplasm, despite being a resident of the mitochondria. Our research has revealed the high-order structural dynamics of Ald4p, showing that adding its substrate, acetaldehyde, can induce its polymerization. By introducing specific mutations that disrupt its catalytic activity, we have been able to show that the formation of these structures is composed of active enzyme. This finding highlights a new paradigm where mitochondrial enzymes can exist and function outside their canonical location, with their polymerization being directly linked to their catalytic state.
Through these studies, we’ve demonstrated that this process of enzyme organization is a widespread phenomenon, not just limited to a few specific proteins or cellular compartments. Ultimately, our goal is to understand how the physical organization of enzymes influences the way cells function, which could have significant implications for medicine and using yeast to model human diseases.
1.Chan C, Sirinonthanawech N, Sato BK, Wilhelm JE, Noree C. Saccharomyces cerevisiae malate dehydrogenase Mdh1p lacking mitochondrial targeting signal can be re-localized to peroxisomes. Biology Open. 2025 Sep 25;14(9):bio062199.
2.Nasalingkhan C, Sirinonthanawech N, Sato BK, Wilhelm JE, Noree C. Localization and regulation of yeast aldehyde dehydrogenase Ald4p structures. Heliyon. 2024 Oct 9;10(20):e39048.
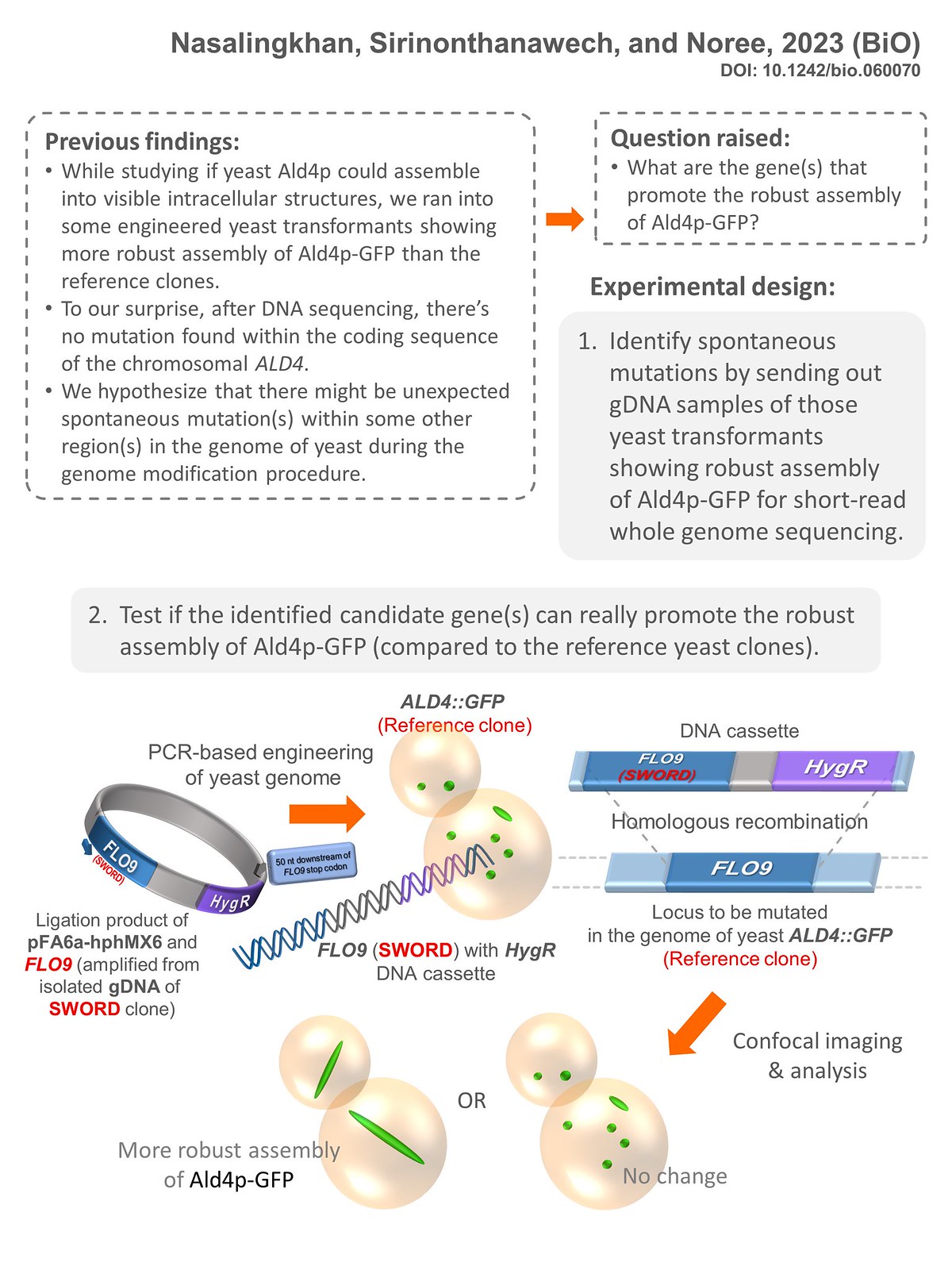
3.Nasalingkhan C, Sirinonthanawech N, Noree C. Robust assembly of the aldehyde dehydrogenase Ald4p in Saccharomyces cerevisiae. Biology Open. 2023 Oct 15;12(10):bio060070.
4.Surasiang T, Noree C. Effects of A6E mutation on protein expression and supramolecular assembly of yeast asparagine synthetase. Biology. 2021 Apr 3;10(4):294.
5.Noree C, Sirinonthanawech N. Nuclear targeted Saccharomyces cerevisiae asparagine synthetases associate with the mitotic spindle regardless of their enzymatic activity. PLoS One. 2020 Dec 21;15(12):e0243742.
6.Noree C, Sirinonthanawech N. Coupled regulations of enzymatic activity and structure formation of aldehyde dehydrogenase Ald4p. Biology Open. 2020 Apr 28;9(4):bio051110.
7.Noree C, Begovich K, Samilo D, Broyer R, Monfort E, Wilhelm JE. A quantitative screen for metabolic enzyme structures reveals patterns of assembly across the yeast metabolic network. Molecular Biology of the Cell. 2019 Oct 1;30(21):2721-2736.
8.Noree C, Sirinonthanawech N, Wilhelm JE. Saccharomyces cerevisiae ASN1 and ASN2 are asparagine synthetase paralogs that have diverged in their ability to polymerize in response to nutrient stress. Scientific Reports. 2019 Jan 22;9(1):278.
9.Noree C, Monfort E, Shotelersuk V. Human asparagine synthetase associates with the mitotic spindle. Biology Open. 2018 Dec 14;7(12):bio038307.
10.Noree C. Extramitochondrial assembly of mitochondrial targeting signal disrupted mitochondrial enzyme aldehyde dehydrogenase. Scientific Reports. 2018 Apr 18;8(1):6186.










Publications and Citations: Google Scholar
Personal Website: norita251.wordpress.com
Academic Tree: Academictree.org








![]()






