Duangrudee Tanramluk creates AI systems to assist in drug design using structural bioinformatics.
Tel: 66 (0) 2441-9003 – 7 Ext. 1211
Email: duangrudee.tan mahidol.ac.th
mahidol.ac.th
Ph.D. (Biochemistry), University of Cambridge, 2010
Academic Program(s)
Molecular Genetics and Genetic Engineering
Molecular and Integrative Biosciences
Systems Biosciences
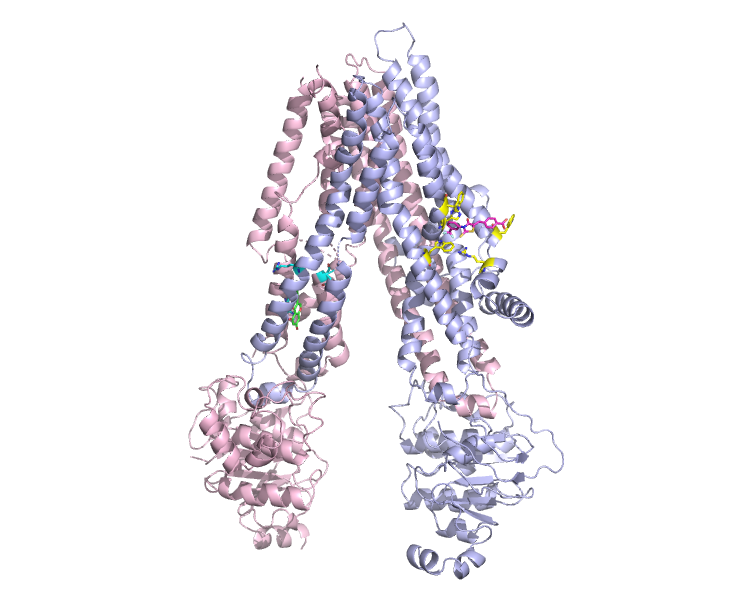
My research experiences, ranging from quantum chemical calculations, protein X-ray crystallography, and software development, have shaped my vision and expertise on how to design tools to solve small molecule drug discovery problems. I choose scientific problems that have big impact on our scientific knowledge and the entire society. To fulfill this objective, I have identified 3 major protein groups, which are the kinases, DHFR, and proteases. Kinases have been implicated in a lot of signaling pathways and human diseases, such as cancer and inflammation. DHFR can be the target protein for several antimicrobial drugs due to its high inhibitor selectivity. In addition, the Main Protease is a major drug target for SARS-CoV-2 antiviral pills.
The research goal is to be able to come up with algorithms that allow for binding affinity prediction to facilitate rational drug design from protein structure coordinates. We are working towards understanding of chemical properties in a rational and interpretable way for inhibitor design. Based on trends of large amount of ligand interactomic distances, shape, and charge complementary in the homologous pockets from structure ensemble, we can now dissect the binding affinity in a human interpretable way for these enzymes.
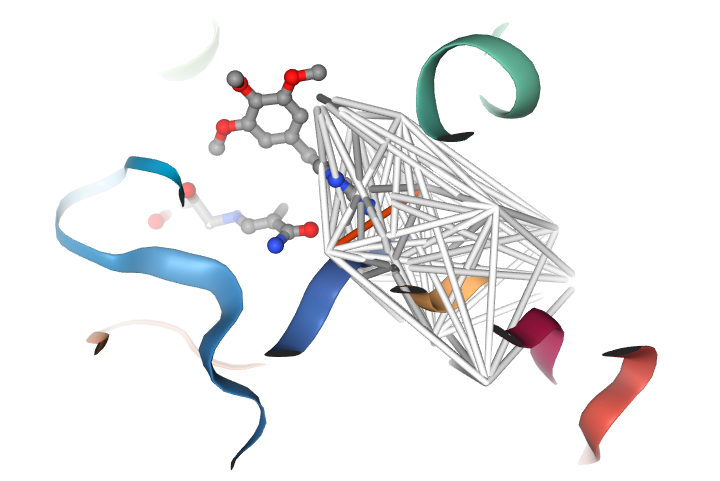
My key inventions are around the webservice Manoraa.org systems to assist drug discovery by linking ligand to target proteins, baseline expression, SNPs, pathways, tissues, and organs as featured in Nucleic Acids Research and Structure. By linking these information in this Manoraa ligand design hub, researchers can perform in silico target discovery and ligand design before planing their wet lab experiments. I have also developed algorithms to analyze conserved features obtained from a set of homologous protein crystal structures which can guide inhibitor design. It enables us to compare the shape of the pocket, to observe position specific interactions, and to display chemical interactions in the pocket of the proteins. Several visualization techniques are employed to proposed the three-dimensional pictures of imaginary drug based on Frequently Occurring Atoms as featured in Scientific Reports. The effects from parameters therein, such as the entities that usually participate in hydrogen bond formation, the parts of the pocket that expand or contract upon binding to inhibitors, can be displayed to guide scientists while formulating an inhibitor design strategy.
Taken together, I gear toward novel computational methods to make drug discovery faster, cheaper, and more effective.
1.Tanramluk D*, Pakotiprapha D, Phoochaijaroen S, Chantravisut P, Thampradid S, Vanichtananku J, Narupiyakul L, Akavipat R, Yuvaniyama J (2022) MANORAA: a machine learning platform to guide protein-ligand design by anchors and influential distances. Structure 30(1): 181-189.e185.
2.Rungruangmaitree R, Phoochaijaroen S, Chimprasit A, Saparpakorn P, Pootanakit K, Tanramluk D* (2023) Structural analysis of the coronavirus main protease for the design of pan-variant inhibitors (2023) Scientific Reports 13(1):7055.
3.Rodrat M, Wongdee K, Teerapornpuntakit J, Thongbunchoo J, Tanramluk D, Aeimlapa R, Thammayon N, Thonapan N, Wattano P, Charoenphandhu N (2022) Vasoactive intestinal peptide and cystic fibrosis transmembrane conductance regulator contribute to the transepithelial calcium transport across intestinal epithelium-like Caco-2 monolayer. PLoS One 17(11): e0277096.
4.Tanramluk D*, Narupiyakul L, Akavipat R, Gong S, Charoensawan V (2016) MANORAA (Mapping Analogous Nuclei Onto Residue And Affinity) for identifying protein-ligand fragment interaction, pathways and SNPs. Nucleic Acids Research 44 (W1): W514-W521. (doi:10.1093/nar/gkw314).
5.Tanramluk D, Schreyer A, Pitt WR, Blundell TL (2009) On the origins of enzyme inhibitor selectivity and promiscuity: a case study of protein kinase binding to staurosporine. Chemical Biology & Drug Design 74(1): 16–24.
2023 Science & Sustainability Award, British Council
2022 Learning Innovation for Online Courses Award: Engage Strategies for Online Teaching
2013 PhD Thesis Award from the National Research Council of Thailand
2008 First Prize Poster Award from the Sanger–Cambridge PhD Symposium (SCAMPS’08), Wellcome Trust Sanger Institute, UK
![]()